Selected ramblings of a geospatial tech nerd
posted by on 26 Mar 2017
No, not lazy as in REST :-) … Lazy as in “Lazy evaluation”:
In computer programming, lazy evaluation is the technique of delaying a computation until the result is required.
Take an example raster processing workflow to go from a bunch of tiled, latlong, GeoTiff digital elevation models to a single shaded relief GeoTiff in projected space:
- Merge the tiles together
- Reproject the merged DEM (using bilinear or cubic interpolation)
- Generate the hillshade from the merged DEM
Simple enough to do with GDAL tools on the command line. Here’s the typical, process-as-you-go implementation:
gdal_merge.py -of GTiff -o srtm_merged.tif srtm_12_*.tif
gdalwarp -t_srs epsg:3310 -r bilinear -of GTiff srtm_merged.tif srtm_merged_3310.tif
gdaldem hillshade srtm_merged_3310.tif srtm_merged_3310_shade.tif -of GTiff
Alternately, we can simulate lazy evaluation by using GDAL Virtual Rasters (VRT) to perform the intermediate steps, only outputting the GeoTiff as the final step.
gdalbuildvrt srtm_merged.vrt srtm_12_0*.tif
gdalwarp -t_srs epsg:3310 -r bilinear -of VRT srtm_merged.vrt srtm_merged_3310.vrt
gdaldem hillshade srtm_merged_3310.vrt srtm_merged_3310_shade2.tif -of GTiff
So what’s the advantage to doing it the VRT way? They both produce exactly the same output raster. Lets compare:
| Process-As-You-Go | "Lazy" VRTs | |
|---|---|---|
| Merge (#1) time | 3.1 sec | 0.05 sec |
| Warp (#2) time | 7.3 sec | 0.10 sec |
| Hillshade (#3) time | 10.5 sec | 19.75 sec |
| Total processing time | 20.9 sec | 19.9 sec |
| Intermediate files | 2 tifs | 2 vrts |
| Intermediate file size | 261 MB | 0.005 MB |
The Lazy VRT method delays all the computationally-intensive processing until it is actually required. The intermediate files, instead of containing the raw raster output of the actual computation, are XML files which contain the instructions to get the desired output. This allows GDAL to do all the processing in one step (the final step #3). The total processing time is not significantly different between the two methods but in terms of the productivity of the GIS analyst, the VRT method is superior. Imagine working with datasets 1000x this size with many more steps - having to type the command, wait 2 hours, type the next, etc. would be a waste of human resources versus assembling the instructions into vrts then hitting the final processing step when you leave the office for a long weekend.
Additionaly, the VRT method produces only small intermediate xml files instead of having a potentially huge data management nightmare of shuffling around GB (or TB) of intermediate outputs! Plus those xml files serve as an excellent piece of metadata which describe the exact processing steps which you can refer to later or adapt to different datasets.
So next time you have a multi-step raster workflow, use the GDAL VRTs to your full advantage - you’ll save yourself time and disk space by being lazy.
Sensitivity analysis made (relatively) simple
posted by on 19 Jan 2014Best bang for your analytical buck
As (geo)data scientists, we spend much of our time working with data models that try (with varying degrees of success) to capture some essential truth about the world while still being as simple as possible to provide a useful abstraction. Inevitably, complexity starts to creep into every model and we don’t often stop to assess the value added by that complexity. When working with models that require a large number of parameters and a huge domain of potential inputs that are expensive to collect, it becomes difficult to answer the question:
What parameters of the model are the most sensitive?
In other words, if I am going to spend my resources obtaining/refining data for this model, where should I focus my efforts in order to get the best bang for the buck? If I spend weeks working on deriving a single parameter for the model, I want some assurance that the parameter is critically important to the model’s prediction. The flip-side, of course, is that if a parameter is not that important to the model’s predictive power, I could save some time by perhaps just using some quick-and-dirty approximation.
SALib: a python module for testing model sensitivity
I was thrilled to find SALib which implements a number of vetted methods for quantitatively assessing parameter sensitivity. There are three basic steps to running SALib:
- Define the parameters to test, define their domain of possible values and generate n sets of randomized input parameters.
- Run the model n times and capture the results.
- Analyze the results to identify the most/least sensitive parameters.
I’ll leave the details of these steps to the SALib documentation. The beauty of the SALib approach is that you have the flexibility[1] to run any model in any way you want, so long as you can manipulate the inputs and outputs adequately.
Case Study: Climate effects on forestry
I wanted to compare a forest growth and yield model under different climate change scenarios in order to assess what the most sensitive climate-related variables were. I identified 4 variables:
- Climate model (4 global circulation models)
- Representative Concentration Pathways (RCPs; 3 different emission trajectories)
- Mortality factor for species viability (0 to 1)
- Mortality factor for equivalent elevation change (0 to 1)
In this case, I was using the Forest Vegetation Simulator(FVS) which requires a configuration file for every model iteration. So, for Step 2, I had to iterate through each set of input variables and use them to generate an appropriate configuration file. This involved translating the real numbers from the samples into categorical variables in some cases. Finally, in order to get the result of the model iteration, I had to parse the outputs of FVS and do some post-processing to obtain the variable of interest (the average volume of standing timber over 100 years). So the flexibility of SALib comes at a slight cost: unless your model works directly with the file formatted for SALib, the input and outputs may require some data manipulation.
After running the all required iterations of the model[2] I was able to analyze the results and assess the sensitivity of the four parameters.
Here’s the output of SALib’s analysis (formatted slightly for readability):
Parameter First_Order First_Order_Conf Total_Order Total_Order_Conf
circulation 0.193685 0.041254 0.477032 0.034803
rcp 0.517451 0.047054 0.783094 0.049091
mortviab -0.007791 0.006993 0.013050 0.007081
mortelev -0.005971 0.005510 0.007162 0.006693
The first order effects represent the effect of that parameter alone. The total order effects are arguably more relevant to understanding the overall interaction of that parameter with your model. The “Conf” columns represent confidence and can be interpreted as error bars.
In this case, we interpret the output as follows:
Parameter Total Order Effect
circulation 0.47 +- 0.03 (moderate influence)
rcp 0.78 +- 0.05 (dominant parameter)
mortviab 0.01 +- 0.007 (weak influence)
mortelev 0.007 +- 0.006 (weak influence)
We can graph each of the input parameters against the results to visualize this:
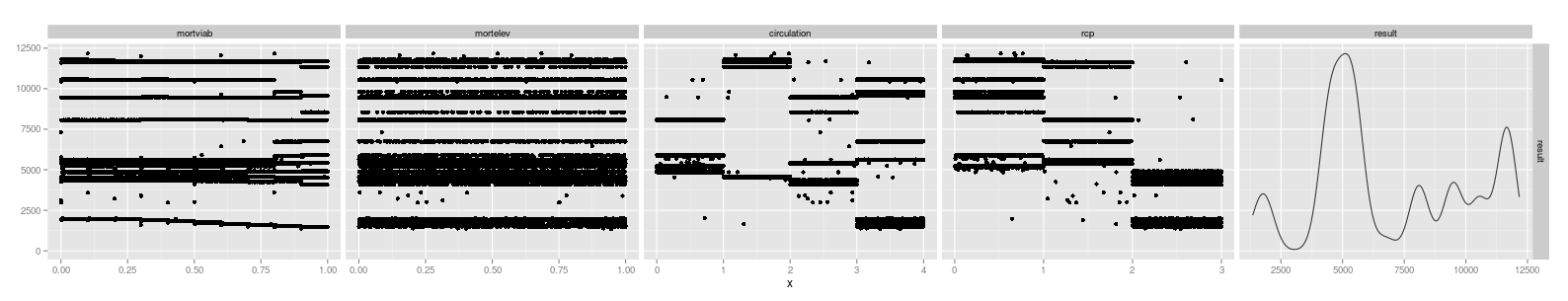
Note that the ‘mortelev’ component is basically flat (as the factor increases, the result stays the same) whereas the choice of ‘rcp’ has a heavy influence (as emissions increase to the highest level, the resulting prediction for timber volumes are noticeably decreased).
The conclusion is that the climate variables, particularly the RCPs related to human-caused emissions, were the strongest determinants[1] of tree growth for this particular forest stand. This ran counter to our initial intuition that the mortality factors would play a large role in the model. Based on this sensitivity analysis, we may be able to avoid wasting effort on refining parameters that are of minor consequence to the output.
Footnotes:
- Compared to more tightly integrated, model-specific methods of sensitivity analysis
- 20 thousand iterations took approximately 8 hours; sensitivity analysis generally requires lots of processing
- Note that the influence of a parameter says nothing about direct causality
Leaflet Simple CSV
posted by on 30 Sep 2013Simple leaftlet-based template for mapping tabular point data on a slippy map
Anyone who’s worked with spatial data and the web has run across the need to take some simple tabular data and put points on an interactive map. It’s the fundamental “Hello World” of web mapping. Yet I always find myself spending way too much time solving this seemingly simple problem. When you consider zoom levels, attributes, interactivity, clustering, querying, etc… it becomes apparent that interactive maps require a bit more legwork. But that functionality is fairly consistent case-to-case so I’ve developed a generalized solution that works for the majority of basic use cases out there:
The idea is pretty generic but useful for most point marker maps:
- Data is in tabular delimited-text (csv, etc.) with two required columns:
latandlng - Points are plotted on full-screen Leaflet map
- Point markers are clustered dynamically based on zoom level.
- Clicking on a point cluster will zoom into the extent of the underlying features.
- Hovering on the point will display the name.
- Clicking will display a popup with columns/properties displayed as an html table.
- Full text filtering with typeahead
- Completely client-side javascript with all dependencies included or linked via CDN
Of course this is mostly just a packaged version of existing work, namely Leaflet with the geoCSV and markercluster plugins.
Usage
- Grab the leaflet-simple-csv zip file and unzip it to a location accessible through a web server.
- Copy the
config.js.templatetoconfig.js - Visit the index.html page to confirm everything is working with the built-in example.
- Customize your
config.jsfor your dataset.
An example config:
var dataUrl = 'data/data.csv';
var maxZoom = 9;
var fieldSeparator = '|';
var baseUrl = 'http://otile{s}.mqcdn.com/tiles/1.0.0/osm/{z}/{x}/{y}.jpg';
var baseAttribution = 'Data, imagery and map information provided by <a href="http://open.mapquest.co.uk" target="_blank">MapQuest</a>, <a href="http://www.openstreetmap.org/" target="_blank">OpenStreetMap</a> and contributors, <a href="http://creativecommons.org/licenses/by-sa/2.0/" target="_blank">CC-BY-SA</a>';
var subdomains = '1234';
var clusterOptions = {showCoverageOnHover: false, maxClusterRadius: 50};
var labelColumn = "Name";
var opacity = 1.0;
The example dataset:
Country|Name|lat|lng|Altitude
United States|New York City|40.7142691|-74.0059738|2.0
United States|Los Angeles|34.0522342|-118.2436829|115.0
United States|Chicago|41.8500330|-87.6500549|181.0
United States|Houston|29.7632836|-95.3632736|15.0
...
I make no claims that this is the “right” way to do it but leveraging 100% client-side javascript libraries and native delimited-text formats seems like the simplest approach. Many of the features included here (clustering, filtering) are useful enough to apply to most situations and hopefully you’ll find it useful.
Python raster stats
posted by on 24 Sep 2013A common task in many of my data workflows involves summarizing geospatial raster datasets based on vector geometries (i.e. zonal statistics). Despite many alternatives (starspan, the QGIS Zonal Statistics plugin, ArcPy and R) there were none that were
- open source
- fast enough
- flexible enough
- worked with python data structures
We’d written a wrapper around starspan for madrona (see madrona.raster_stats ) but relying on shell calls and an aging, unmaintained C++ code base was not cutting it.
So I set out to create a solution using numpy, GDAL and python. The
rasterstats package was born.
`python-raster-stats` on github
Example
Let’s jump into an example. I’ve got a polygon shapefile of continental US state boundaries and a raster dataset of annual precipitation from the North American Environmental Atlas.
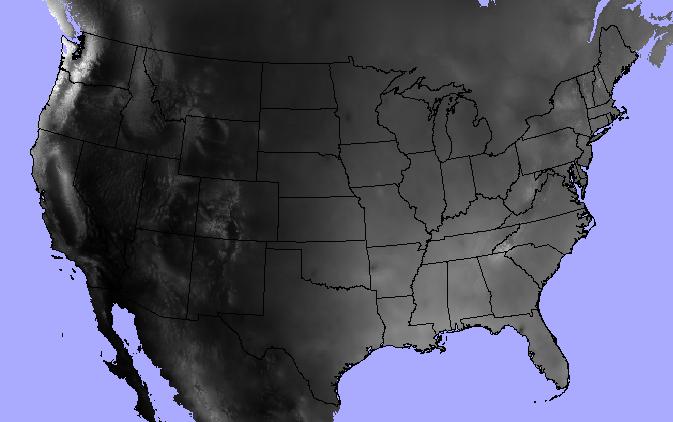
>>> states = "/data/workspace/rasterstats_blog/boundaries_contus.shp"
>>> precip = "/data/workspace/rasterstats_blog/NA_Annual_Precipitation_GRID/NA_Annual_Precipitation/data/na_anprecip/hdr.adf"
The raster_stats function is the main entry point. Provide a vector and a
raster as input and expect a list of dicts, one for each input feature.
>>> from rasterstats import raster_stats
>>> rain_stats = raster_stats(states, precip, stats="*", copy_properties=True)
>>> len(rain_stats) # continental US; 48 states plus District of Columbia
49
Print out the stats for a given state:
>>> [x for x in rain_stats if x['NAME'] == "Oregon"][0]
{'COUNTRY': 'USA',
'EDIT': 'NEW',
'EDIT_DATE': '20060803',
'NAME': 'Oregon',
'STATEABB': 'US-OR',
'Shape_Area': 250563567264.0,
'Shape_Leng': 2366783.00361,
'UIDENT': 124704,
'__fid__': 35,
'count': 250510,
'majority': 263,
'max': 3193.0,
'mean': 779.2223903237395,
'median': 461.0,
'min': 205.0,
'minority': 3193,
'range': 2988.0,
'std': 631.539502512283,
'sum': 195203001.0,
'unique': 2865}
Find the three driest states:
>>> [(x['NAME'], x['mean']) for x in
sorted(rain_stats, key=lambda k: k['mean'])[:3]]
[('Nevada', 248.23814034118908),
('Utah', 317.668743027571),
('Arizona', 320.6157232064074)]
And write the data out to a csv.
from rasterstats import stats_to_csv
with open('out.csv', 'w') as fh:
fh.write(stats_to_csv(rain_stats))
Geo interface
The basic usage above shows the path of an entire OGR vector layer as the first argument. But raster-stats also supports other vector features/geometries.
- Well-Known Text/Binary
- GeoJSON string and mappings
- Any python object that supports the geo_interface
- Single objects or iterables
In this example, I use a geojson-like python mapping to specify a single geometry
>>> geom = {'coordinates': [[
[-594335.108537269, -570957.932799394],
[-422374.54395311, -593387.5716581973],
[-444804.1828119133, -765348.1362423564],
[-631717.839968608, -735441.9510972851],
[-594335.108537269, -570957.932799394]]],
'type': 'Polygon'}
>>> raster_stats(geom, precip, stats="min median max")
[{'__fid__': 0, 'max': 1011.0, 'median': 451.0, 'min': 229.0}]
Categorical
We’re not limited to descriptive statistics for continuous rasters either; we can get unique pixel counts for categorical rasters as well. In this example, we’ve got a raster of 2005 land cover (i.e. general vegetation type).
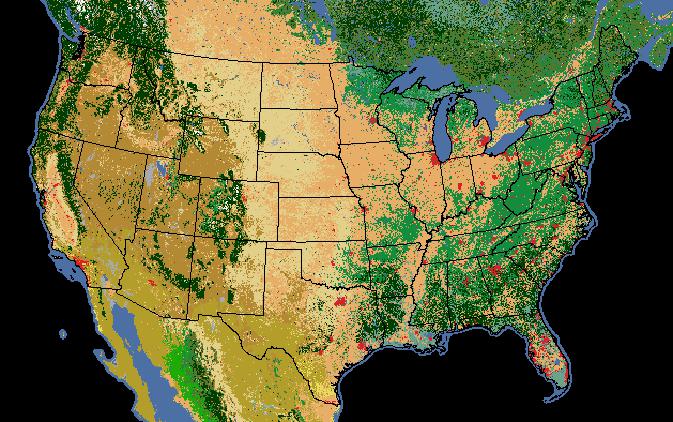
Note that
we can specify only the stats that make sense and the categorical=True
provides a count of each pixel value.
>>> landcover = "/data/workspace/rasterstats_blog/NA_LandCover_2005.img"
>>> veg_stats = raster_stats(states, landcover,
stats="count majority minority unique",
copy_properties=True,
nodata_value=0,
categorical=True)
>>> [x for x in veg_stats if x['NAME'] == "Oregon"][0]
{1: 999956,
3: 6,
5: 3005,
6: 198535,
8: 2270805,
10: 126199,
14: 20883,
15: 301884,
16: 17452,
17: 39246,
18: 28872,
19: 2174,
'COUNTRY': 'USA',
'EDIT': 'NEW',
'EDIT_DATE': '20060803',
'NAME': 'Oregon',
'STATEABB': 'US-OR',
'Shape_Area': 250563567264.0,
'Shape_Leng': 2366783.00361,
'UIDENT': 124704,
'__fid__': 35,
'count': 4009017,
'majority': 8,
'minority': 3,
'unique': 12}
Of course the pixel values alone don’t make much sense. We need to interpret the pixel values as land cover classes:
Value, Class_name
1 Temperate or sub-polar needleleaf forest
2 Sub-polar taiga needleleaf forest
3 Tropical or sub-tropical broadleaf evergreen
4 Tropical or sub-tropical broadleaf deciduous
5 Temperate or sub-polar broadleaf deciduous
6 Mixed Forest
7 Tropical or sub-tropical shrubland
8 Temperate or sub-polar shrubland
9 Tropical or sub-tropical grassland
10 Temperate or sub-polar grassland
11 Sub-polar or polar shrubland-lichen-moss
12 Sub-polar or polar grassland-lichen-moss
13 Sub-polar or polar barren-lichen-moss
14 Wetland
15 Cropland
16 Barren Lands
17 Urban and Built-up
18 Water
19 Snow and Ice
So, for our Oregon example above we can see that, despite Oregon’s reputation as a lush green landscape, the majority land cover class (#8) is “Temperate or sub- polar shrubland” at 2.27m pixels out of 4 millions total.
There’s a lot more functionality that isn’t covered in this post but you get the picture… please check it out and let me know what you think.
Creating UTFGrids directly from a polygon datasource
posted by on 20 Aug 2012We’ve begun to rely on the interactivity provided by UTFGrids in many of our recent web maps. (Quick recap: UTFGrids are “invisible” map tiles that allow direct interactivity with feature attributes without querying the server.) Earlier this year, I created the initial OpenLayers UTFGrid support and was glad to see it accepted into OpenLayer 2.12 (with some enhancements).
With the client-side javascript support in place, the only missing piece in the workflow was to create the UTFGrid .json files. We had expirimented with several alternate UTFGrid renderers but Mapnik’s rendering was by far the fastest and produced the best results. Using Tilemill was a convenient way to leverage the Mapnik UTFGrid renderer but it came at the cost of a somewhat circuitious and manual workflow:
- Load the data up into Tilemill,
- Configure interactivity fields
- Export to .mbtiles
- Convert to .json files
What we really needed was a script to take a polygon shapefile and render the UTFGrids directly to files. Mapnik would provide the rendering while the Global Map Tiles python module would provide the logic for going back and forth between geographic coordinates and tile grid coordinates. From there it’s just a matter of determining the extent of the data set and, for a specified set of zoom levels, looping through and using Mapnik to render the UTFGrid to a .json file in Z/X/Y.json directory structure.
Get `create-utfgrids` on github
If we have a mercator polygon shapefile of ecoregions and want to render UTFGrids for zoom levels 3 through 5 using the dom_desc and div_desc attributes, we could use a command like
$ ./create_utfgrids.py test_data/bailey_merc.shp 3 5 ecoregions -f dom_desc,div_desc
WARNING:
This script assumes a polygon shapefile in spherical mercator projection.
If any of these assumptions are not true, don't count on the results!
* Processing Zoom Level 3
* Processing Zoom Level 4
* Processing Zoom Level 5
and inspect the output (e.g. zoom level 5, X=20, Y=18)
$ cat ecoregions/5/20/18.json | python -mjson.tool
{
"data": {
"192": {
"div_desc": "RAINFOREST REGIME MOUNTAINS",
"dom_desc": "HUMID TROPICAL DOMAIN"
},
...
"grid": [
" !!!!!!!!!#####$%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%",
...
Some caveats:
- This currently only works for polygon datasets in a Web Mercator projection.
- It’s only tested with shapefiles as it assumes a single-layer datasource at the moment. Full OGR Datasource support would not be too difficult to add for PostGIS, etc.
- It assumes a top-origin tile scheme (as do OSM and Google Maps). Supporting TMS bottom-origin schemes in the future should be straightforward.
- Requires OGR and Mapnik >= 2.0 with python bindings. Finding windows binaries for the required version of Mapnik may be difficult so using OSX/Linux is recommended at this time.
Many thanks to Dane Springmeyer for his help on UTFGrid related matters and and to Klokan Petr Přidal for his MapTiler docs
